Naysayers would say that government, and hence taxpayer dollars, should not be used to fund science initiatives. After all academia and business seem to do a fairly good job of discovery and innovation without a helping hand pilfering from the public purse. And, without a doubt, and money aside, government funded projects do raise a number of thorny questions: On what should our hard-earned income tax be spent? Who decides on the priorities? How is progress to be measured? Do taxpayers get any benefit in return? After many of us cringe at the thought of an unelected bureaucrat or a committee of such spending millions if not billions of our dollars. Why not just spend the money on fixing our national potholes?
But despite our many human flaws and foibles we are at heart explorers. We seek to know more about ourselves, our world and our universe. Those who seek answers to fundamental questions of consciousness, aging, and life are pioneers in this quest to expand our domain of understanding and knowledge. These answers increasingly aid our daily lives through continuous improvement in medical science, and innovation in materials science. And, our collective lives are enriched as we increasingly learn more about the how and the why of our and our universe’s existence.
So, some of our dollars have gone towards big science at the Large Hadron Collider (LHC) beneath Switzerland looking for constituents of matter, the wild laser experiment at the National Ignition Facility designed to enable controlled fusion reactions, and the Curiosity rover exploring Mars. Yet more of our dollars have gone to research and development into enhanced radar, graphene for next generation circuitry, online courseware, stress in coral reefs, sensors to aid the elderly, ultra-high speed internet for emergency response, erosion mitigation, self-cleaning surfaces, flexible solar panels.
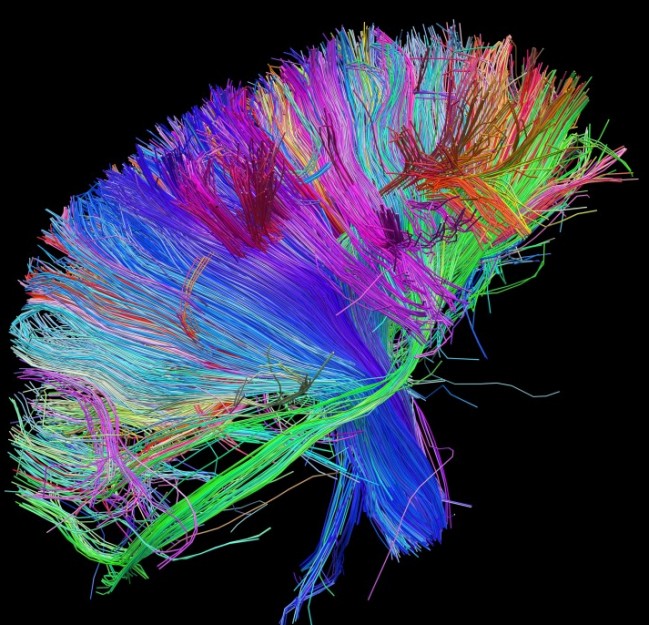
Now comes word that the U.S. government wants to spend $3 billion dollars — over 10 years — on building a comprehensive map of the human brain. The media has dubbed this the “connectome” following similar efforts to map our human DNA, the genome. While this is the type of big science that may yield tangible results and benefits only decades from now, it ignites the passion and curiosity of our children to continue to seek and to find answers. So, this is good news for science and the explorer who lurks within us all.
[div class=attrib]From ars technica:[end-div]
Over the weekend, The New York Times reported that the Obama administration is preparing to launch biology into its first big project post-genome: mapping the activity and processes that power the human brain. The initial report suggested that the project would get roughly $3 billion dollars over 10 years to fund projects that would provide an unprecedented understanding of how the brain operates.
But the report was remarkably short on the scientific details of what the studies would actually accomplish or where the money would actually go. To get a better sense, we talked with Brown University’s John Donoghue, who is one of the academic researchers who has been helping to provide the rationale and direction for the project. Although he couldn’t speak for the administration’s plans, he did describe the outlines of what’s being proposed and why, and he provided a glimpse into what he sees as the project’s benefits.
What are we talking about doing?
We’ve already made great progress in understanding the behavior of individual neurons, and scientists have done some excellent work in studying small populations of them. On the other end of the spectrum, decades of anatomical studies have provided us with a good picture of how different regions of the brain are connected. “There’s a big gap in our knowledge because we don’t know the intermediate scale,” Donaghue told Ars. The goal, he said, “is not a wiring diagram—it’s a functional map, an understanding.”
This would involve a combination of things, including looking at how larger populations of neurons within a single structure coordinate their activity, as well as trying to get a better understanding of how different structures within the brain coordinate their activity. What scale of neuron will we need to study? Donaghue answered that question with one of his own: “At what point does the emergent property come out?” Things like memory and consciousness emerge from the actions of lots of neurons, and we need to capture enough of those to understand the processes that let them emerge. Right now, we don’t really know what that level is. It’s certainly “above 10,” according to Donaghue. “I don’t think we need to study every neuron,” he said. Beyond that, part of the project will focus on what Donaghue called “the big question”—what emerges in the brain at these various scales?”
While he may have called emergence “the big question,” it quickly became clear he had a number of big questions in mind. Neural activity clearly encodes information, and we can record it, but we don’t always understand the code well enough to understand the meaning of our recordings. When I asked Donaghue about this, he said, “This is it! One of the big goals is cracking the code.”
Donaghue was enthused about the idea that the different aspects of the project would feed into each other. “They go hand in hand,” he said. “As we gain more functional information, it’ll inform the connectional map and vice versa.” In the same way, knowing more about neural coding will help us interpret the activity we see, while more detailed recordings of neural activity will make it easier to infer the code.
As we build on these feedbacks to understand more complex examples of the brain’s emergent behaviors, the big picture will emerge. Donaghue hoped that the work will ultimately provide “a way of understanding how you turn thought into action, how you perceive, the nature of the mind, cognition.”
How will we actually do this?
Perception and the nature of the mind have bothered scientists and philosophers for centuries—why should we think we can tackle them now? Donaghue cited three fields that had given him and his collaborators cause for optimism: nanotechnology, synthetic biology, and optical tracers. We’ve now reached the point where, thanks to advances in nanotechnology, we’re able to produce much larger arrays of electrodes with fine control over their shape, allowing us to monitor much larger populations of neurons at the same time. On a larger scale, chemical tracers can now register the activity of large populations of neurons through flashes of fluorescence, giving us a way of monitoring huge populations of cells. And Donaghue suggested that it might be possible to use synthetic biology to translate neural activity into a permanent record of a cell’s activity (perhaps stored in DNA itself) for later retrieval.
Right now, in Donaghue’s view, the problem is that the people developing these technologies and the neuroscience community aren’t talking enough. Biologists don’t know enough about the tools already out there, and the materials scientists aren’t getting feedback from them on ways to make their tools more useful.
Since the problem is understanding the activity of the brain at the level of large populations of neurons, the goal will be to develop the tools needed to do so and to make sure they are widely adopted by the bioscience community. Each of these approaches is limited in various ways, so it will be important to use all of them and to continue the technology development.
Assuming the information can be recorded, it will generate huge amounts of data, which will need to be shared in order to have the intended impact. And we’ll need to be able to perform pattern recognition across these vast datasets in order to identify correlations in activity among different populations of neurons. So there will be a heavy computational component as well.
[div class=attrib]Read the entire article following the jump.[end-div]
[div class=attrib]Image: White matter fiber architecture of the human brain. Courtesy of the Human Connectome Project.[end-div]